Computational Tools
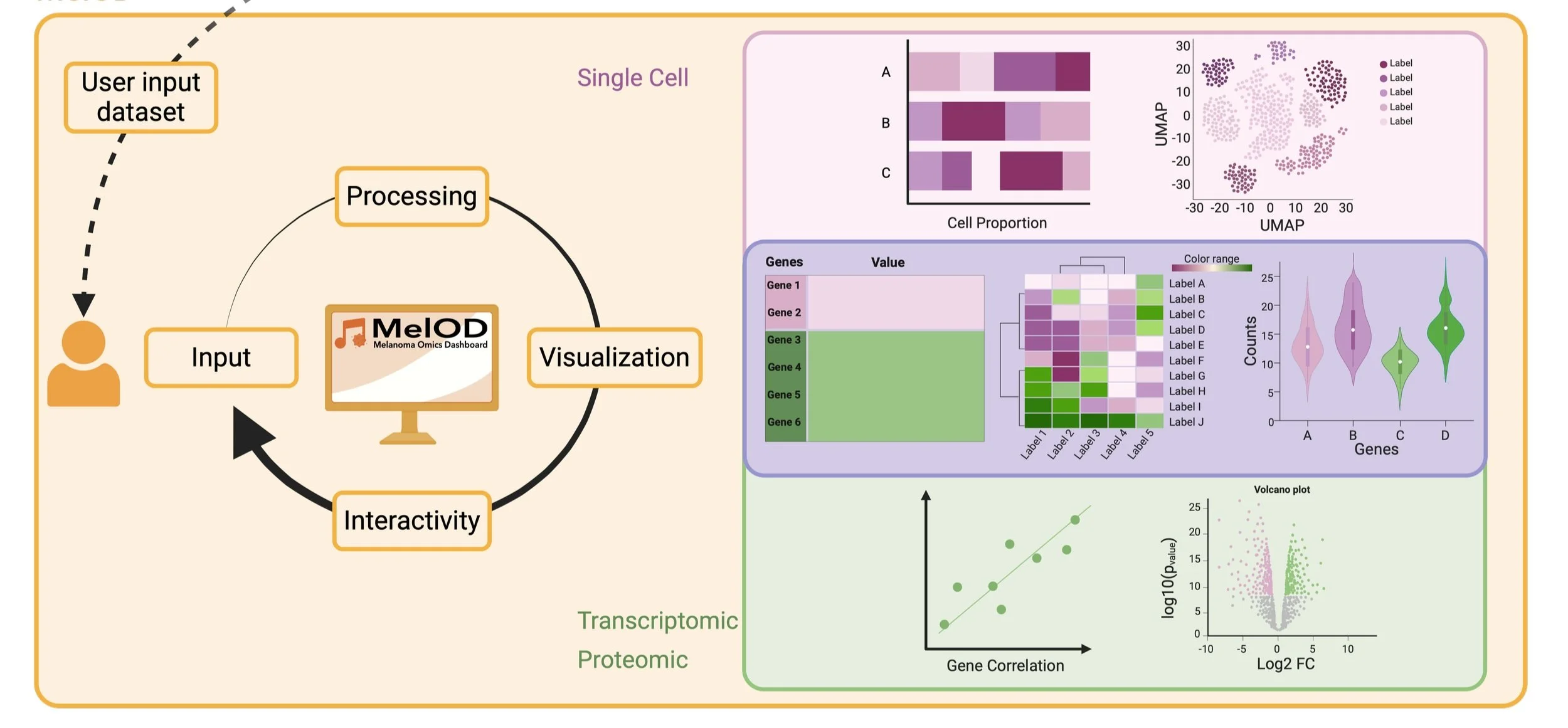
Melanoma Omics Dashboard (melOD)
https://ruggleslab.shinyapps.io/MelOD/
An interactive RShiny application that streamlines the exploration and visualization of melanoma-specific omics data, including transcriptomics, single-cell RNA-seq, and proteomics. It provides tools for differential expression, survival analysis, and gene expression dynamics within a user-friendly interface.
Repository: https://github.com/ruggleslab/MelOD
UbiDash
https://ruggleslab.shinyapps.io/UbiDash/
A web interface for querying and generating interactive plots from the CPTAC, CCLE, and PRIDE datasets. It supports oncoplots, lollipop plots, volcano and boxplots for mutation analysis, as well as customizable heatmaps, volcano plots, density plots, and correlation plots for expression data.
Repository: https://github.com/ruggleslab/ubiDash
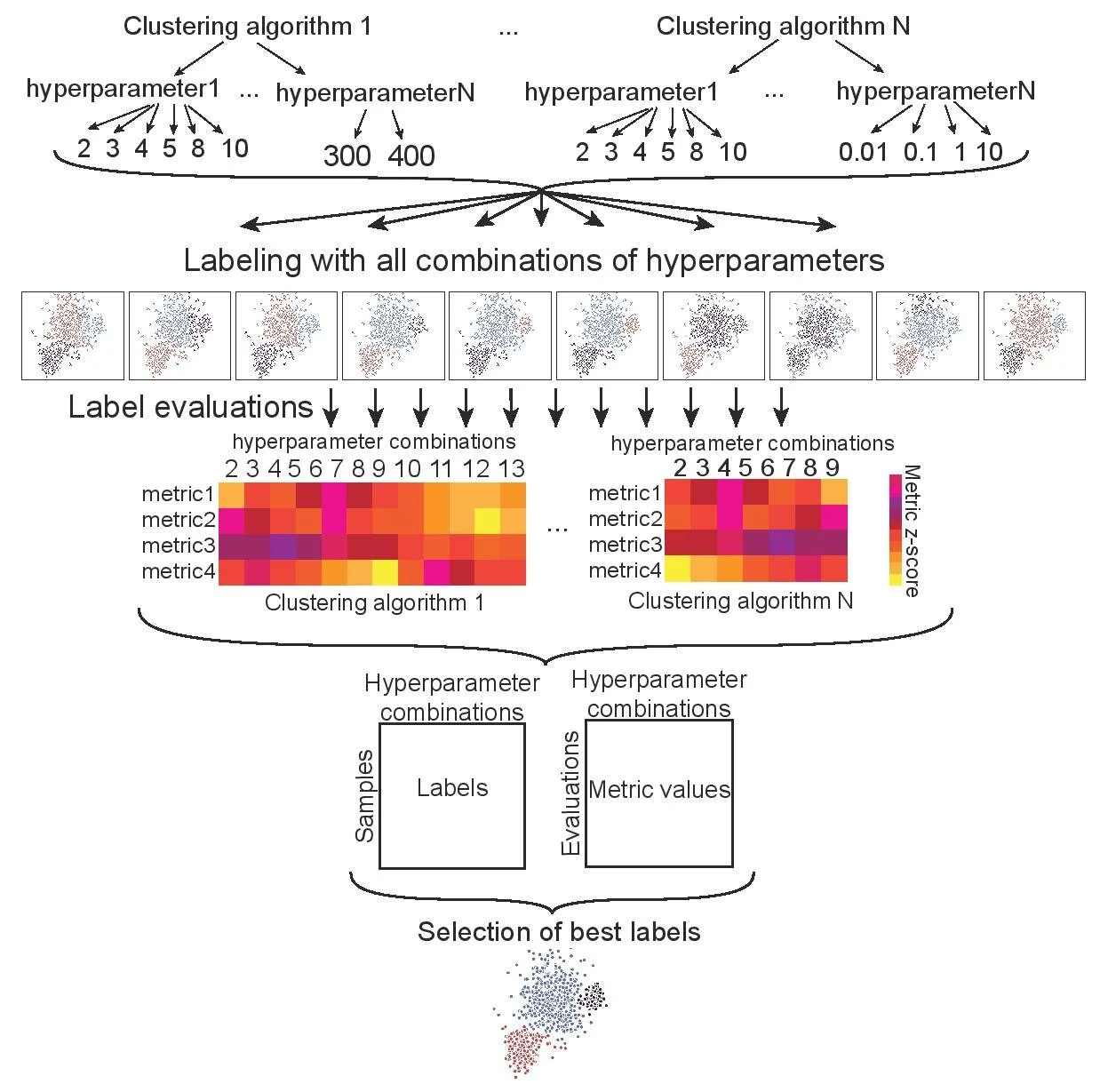
HyperCluster
A python package and SnakeMake workflow for parallelized clustering optimization.
Github: https://github.com/liliblu/hypercluster
Documentation: https://hypercluster.readthedocs.io/en/latest/
Publication:
Blumenberg L, Ruggles KV. (2020) Hypercluster: a flexible tool for parallelized unsupervised clustering optimization BMC Bioinformatics
BlackSheep
A tool for differential extreme-value analysis with packages in both Python and R.
Python version:
R version:
Publication:
Blumenberg L, Kawaler E., Cornwell M, Smith S, Ruggles KV, Fenyo D. (2019) BlackSheep: A Bioconductor and Bioconda package for differential extreme value analysis. bioRxiv 825067
Workflow Hub for Automated Metagenomic Exploration (WHAM!)
Rshiny web-tool allowing for exploratory analysis of metagenomics and metatranscriptomic data. Includes visualization and statistical analysis on the gene family and taxa level.
Publication:
Devlin JC, Battaglia T, Blaser MJ, Ruggles KV. (2018) WHAM!: a web-based visualization suite for user-defined analysis of metagenomic shotgun sequence data. BMC Genomics. PMID: 29940835
Ribosomal Investigation and Visualization to Evaluate Translation (RIVET)
A simple to use tool to automate the statistical analysis of RNA seq data acquired from polysome profile or ribosome footprinting experiments.
Publication:
Ernlund AE, Schnieder RJ, Ruggles KV. (2018) RIVET: comprehensive graphic user interface for analysis and exploration of genome-wide translatomics data. BMC Genomics 19:809