Computational Tools
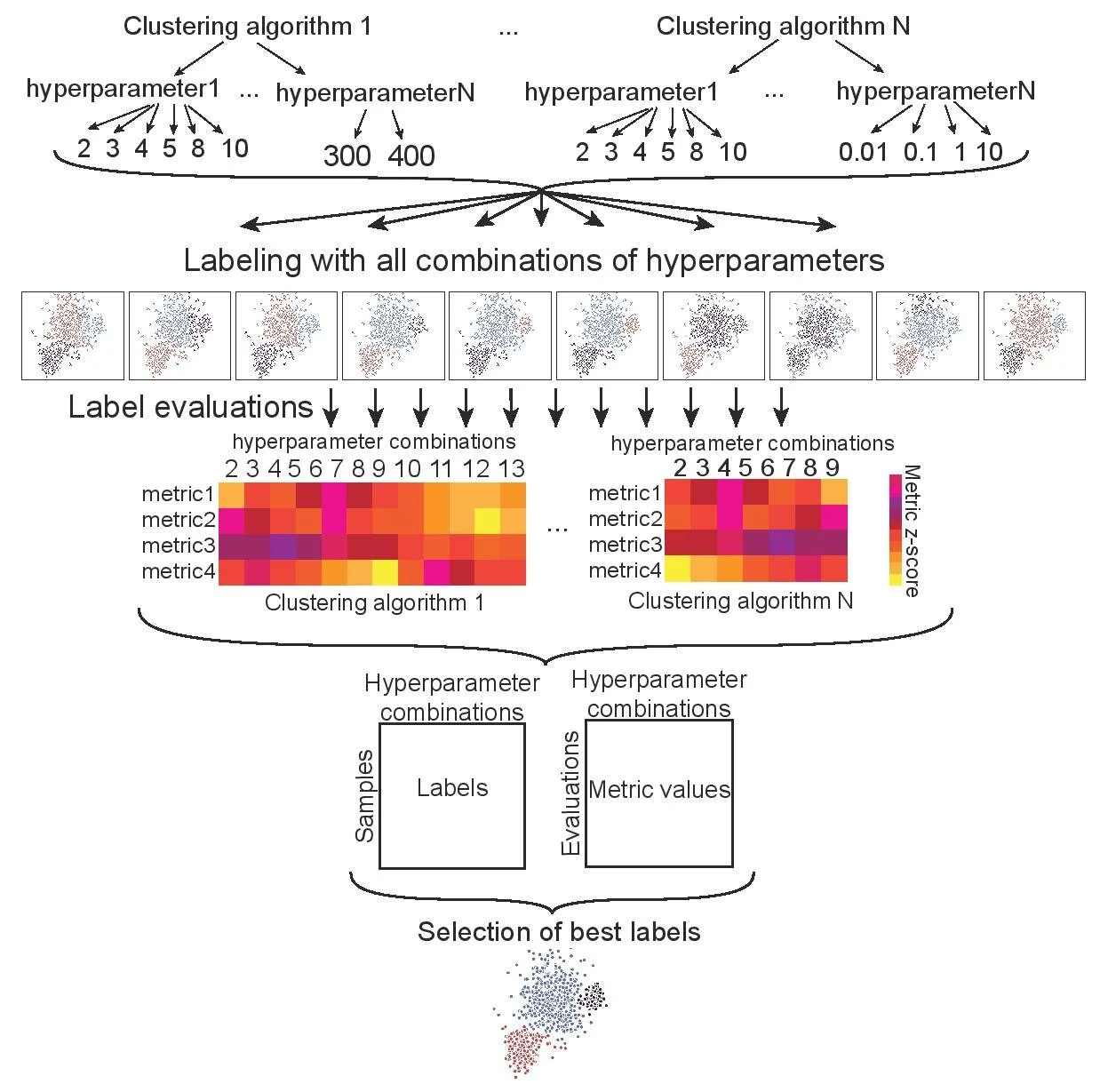
HyperCluster
A python package and SnakeMake workflow for parallelized clustering optimization.
Github: https://github.com/liliblu/hypercluster
Documentation: https://hypercluster.readthedocs.io/en/latest/
Publication:
Blumenberg L, Ruggles KV. (2020) Hypercluster: a flexible tool for parallelized unsupervised clustering optimization BMC Bioinformatics
BlackSheep
A tool for differential extreme-value analysis with packages in both Python and R.
Python version:
R version:
Publication:
Blumenberg L, Kawaler E., Cornwell M, Smith S, Ruggles KV, Fenyo D. (2019) BlackSheep: A Bioconductor and Bioconda package for differential extreme value analysis. bioRxiv 825067
Workflow Hub for Automated Metagenomic Exploration (WHAM!)
Rshiny web-tool allowing for exploratory analysis of metagenomics and metatranscriptomic data. Includes visualization and statistical analysis on the gene family and taxa level.
Publication:
Devlin JC, Battaglia T, Blaser MJ, Ruggles KV. (2018) WHAM!: a web-based visualization suite for user-defined analysis of metagenomic shotgun sequence data. BMC Genomics. PMID: 29940835
Ribosomal Investigation and Visualization to Evaluate Translation (RIVET)
A simple to use tool to automate the statistical analysis of RNA seq data acquired from polysome profile or ribosome footprinting experiments.
Publication:
Ernlund AE, Schnieder RJ, Ruggles KV. (2018) RIVET: comprehensive graphic user interface for analysis and exploration of genome-wide translatomics data. BMC Genomics 19:809